Research
Protein-nucleic acid interactions
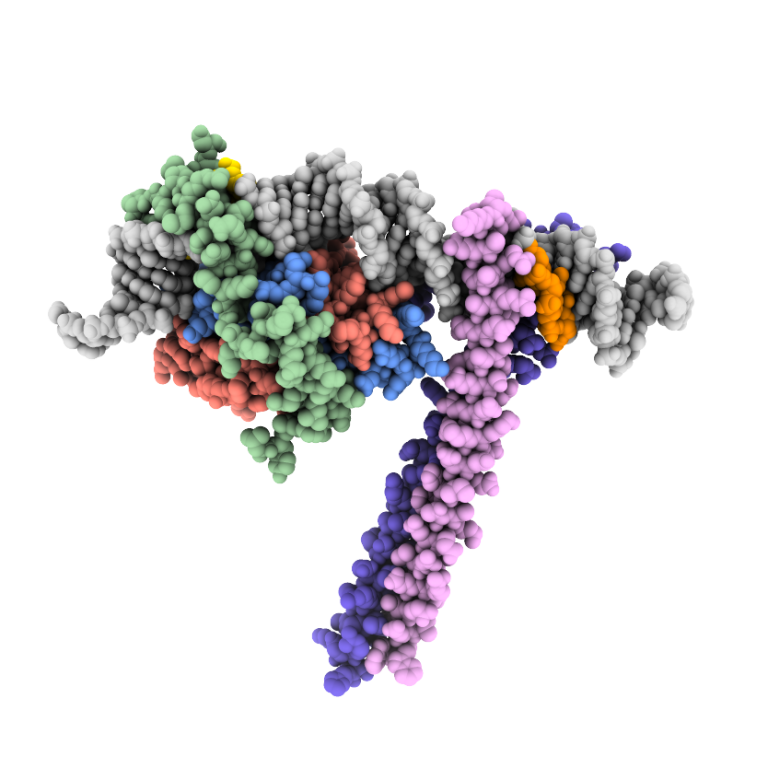
Protein-nucleic acid complexes are fundamental to life. We investigate structures of transcription factors and tRNA-modifying enzymes.
Proteasome biology
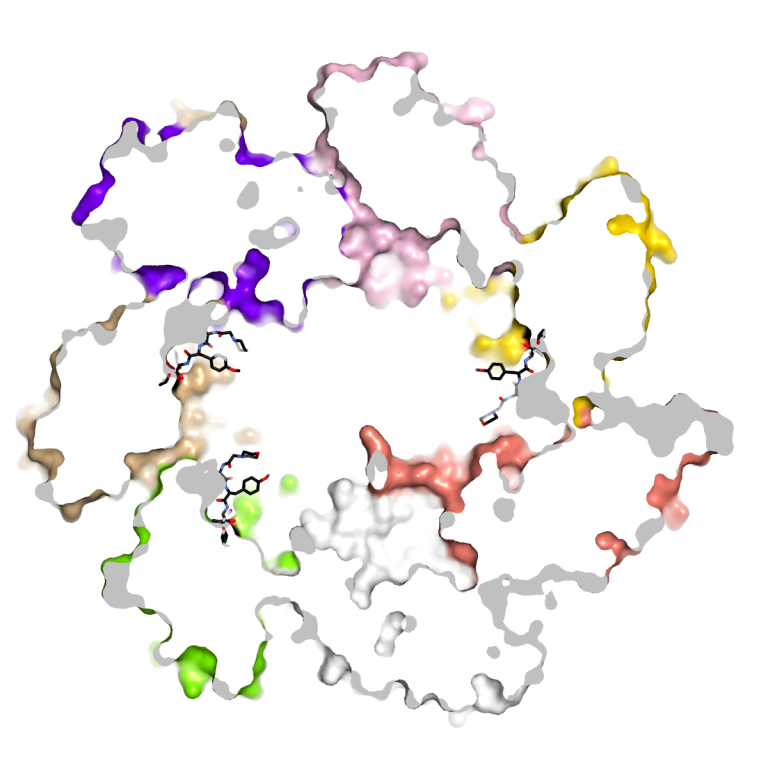
20S proteasomes are multisubunit proteases. Studying their structural features is of interest for diverse biological settings and diseases.
Natural product
biosynthesis
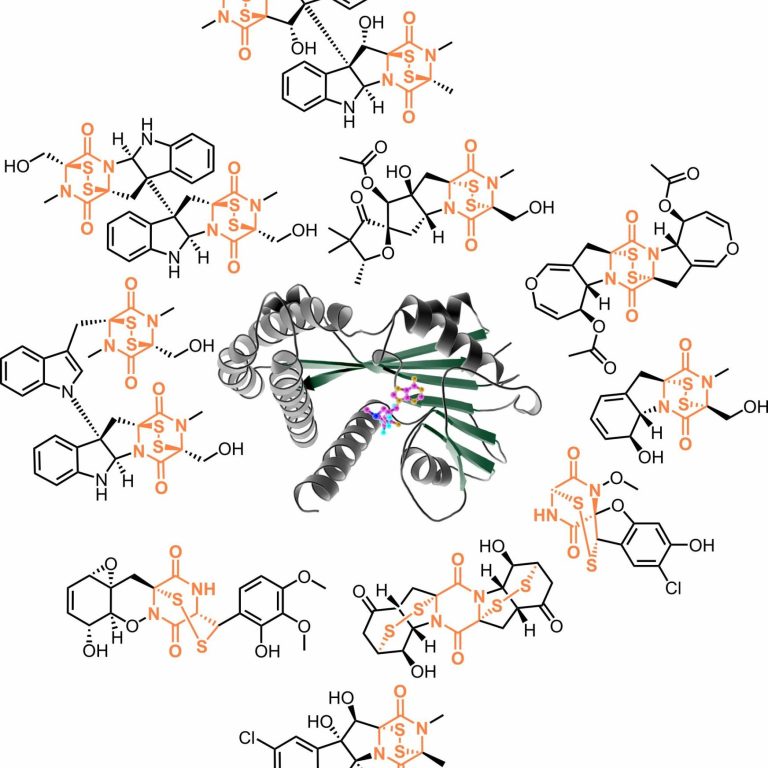
Enzymes are selective and efficient catalysts. By visualizing how enzymes bind their ligands, we learn about their reaction mechanism.
We predominantly use X-ray crystallography and a set of biochemical tools to study proteins with respect to their structure and function.
Protein-nucleic acid interactions
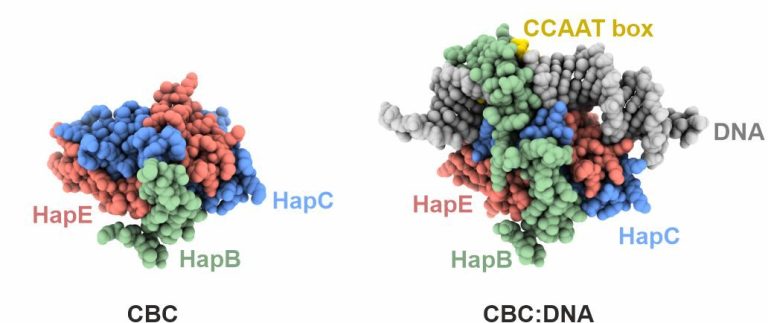
The CCAAT box is a ubiquitous element of eukaryotic promoters. It is specifically bound by the heterotrimeric CCAAT-binding complex (CBC) which acts as a master regulator to control transcriptional activation and repression. In collaboration with the groups of Prof. Dr. Axel Brakhage in Jena and Prof. Dr. Hubertus Haas in Innsbruck, we investigate the structure and function of the CBC in fungi.
1. Structure of the core complex
Initial work characterized the structure of the core complex alone and bound to double stranded DNA. The subunits HapC and HapE adopt a histone fold and bend the target DNA in a nucleosome-like manner. The third subunit, HapB, interacts with HapC as well as HapE and directly contacts the CCAAT motif by inserting into the minor groove. The crystallographic data explained how the CBC gains sequence specificity for the CCAAT box.
Huber, E. M., Scharf, D. H., Hortschansky, P., Groll, M., Brakhage, A. A.
DNA minor groove sensing and widening by the CCAAT-binding complex
Structure, 2012, 20 (10), 1757-68
2. Investigation of a drug resistance mechanism
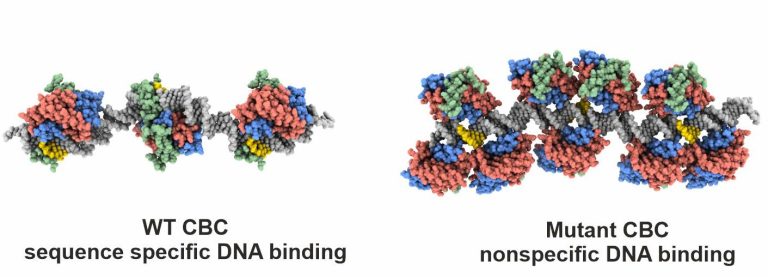
Azoles are commonly used to treat human or plant fungal infections, but resistances are on the rise. We therefore investigated how the single point mutation P88L in subunit HapE of the CBC provokes fungal azole resistance. Our X-ray structure visualized that the mutant HapE subunit interferes with the CBC’s function to bend and properly recognize its target DNA sequence, leading to transcriptional derepression of the cyp51A gene and overproduction of the 14-α sterol demethylase Cyp51A, the target of azoles.
Hortschansky, P., Misslinger, M., Mörl, Y., Gsaller, F., Bromley, M. J., Brakhage, A. A., Groll, M., Haas, H., Huber, E. M.
Structural basis of HapEP88L-linked antifungal triazole resistance in Aspergillus fumigatus
Life Sci. Alliance, 2020, 3 (7), e202000729, 1-12
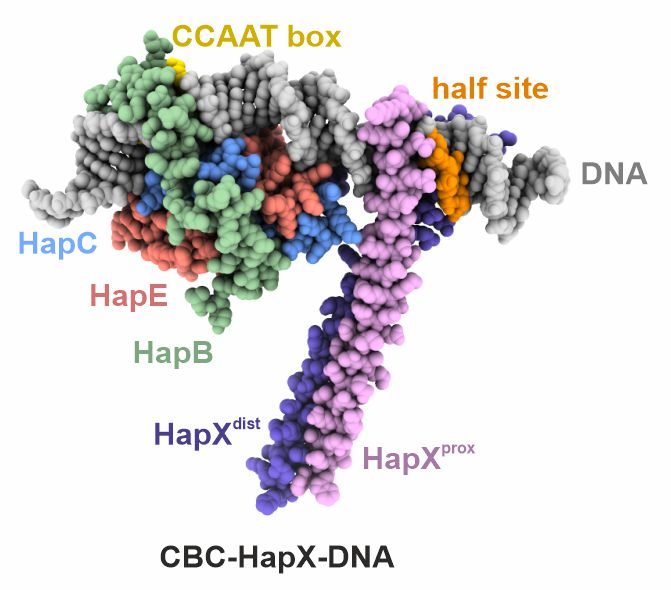
3. Cooperativity with other transcription factors
In certain fungi the CBC teams up with the dimeric basic leucine zipper HapX to control transcription of genes involved in iron metabolism. Despite detailed biochemical data, the structural basis of cooperativity between CBC and HapX remained unclear. Our crystal structure of the CBC-HapX-DNA complex now illustrates how the five-subunit assembly mediates recognition of three separate DNA motifs and how asymmetry is used to establish a 2:1 interaction between a homodimer and its binding partner.
Huber, E. M., Hortschansky, P., Scheven, M. T., Misslinger, M., Haas, H., Brakhage, A. A., Groll, M.
Structural insights into cooperative DNA recognition by the CCAAT-binding complex and its bZIP transcription factor HapX
Structure, 2022, 30(7), 934-946
tRNA modifying enzymes
Structural basis for transcriptional regulation in fungi
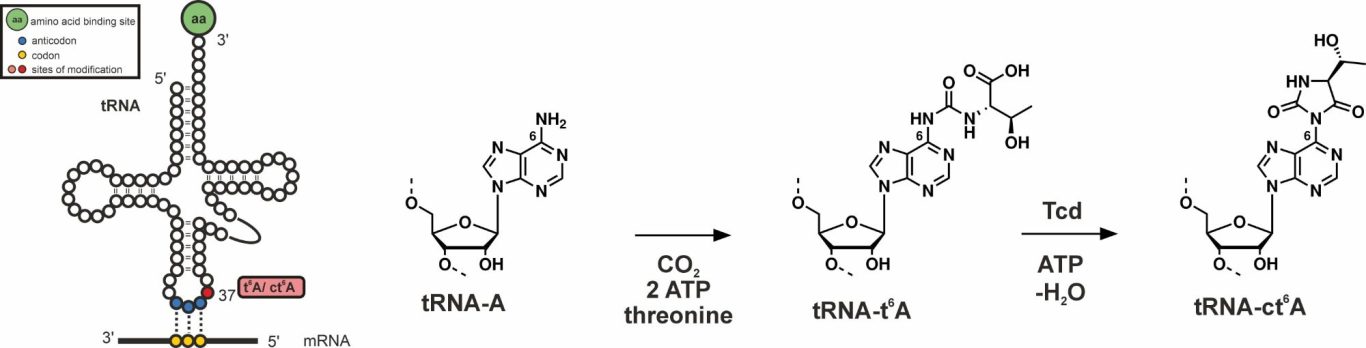
tRNAs are decorated with numerous base modifications to stabilize their structure and to assist their decoding function at the ribosome. tRNAs that decode ANN base triplets in mRNAs often bear threonylcarbamoyladenosine (t6A) at position 37 near the anticodon stem loop. While this modification is essential and universally conserved, some bacteria, fungi, plants and protists have evolved a cyclic non-essential version, termed ct6A. While the enzymatic formation of t6A from L-threonine, ATP and bicarbonate is well understood, the subsequent cyclization reaction to ct6A by threonylcarbamoyladenosine dehydratases (Tcd enzymes) remains elusive. We investigate the molecular mechanisms and chemical reactions by which Tcd enzymes post-transcriptionally install the epigenetic modification ct6A on tRNA.
Natural product biosynthesis
Structural aspects of epidithiodioxopiperazine biosynthesis
Epidithiodioxopiperazines (ETPs) are fungal secondary metabolites with diverse biological activities. Their 2,5-diketopiperazine (DKP) scaffold, built from two amino acids, is bridged by a reactive disulfur moiety that is essential for bioactivity. Gliotoxin is the most prominent member of this growing group of natural sulfur compounds. We study enzymes involved in the biosynthesis of gliotoxin or related ETPs and DKPs from a structural and biochemical perspective. X-ray structures of key biosynthesis enzymes with ligands provide mechanistic insights.

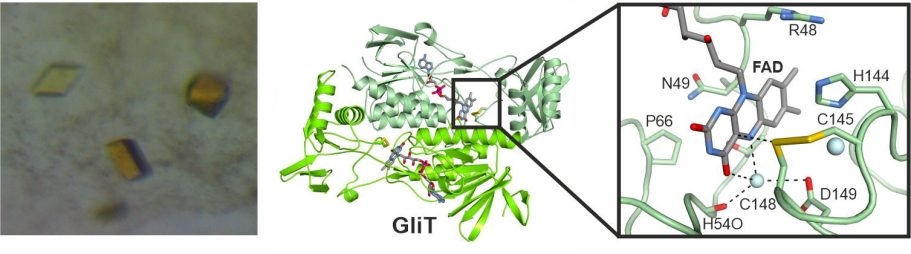
In 2014, we reported the crystal structure of the flavin adenine dinucleotide-dependent oxidoreductase GliT, which installs the disulfide bridge in gliotoxin. Together with data on two related enzymes, a unifying reaction mechanism was proposed. Using mutants of GliT and related enzymes, we aim for trapping a reaction intermediate in the enzymes' active site.
Scharf, D. H., Groll, M., Habel, A., Heinekamp, T., Hertweck, C., Brakhage, A. A., Huber, E. M.
Flavoenzyme-catalyzed disulfide bridge formation in natural products
Angew. Chem. Int. Ed., 2014, 53 (8), 2221–4
In the following, we also analyzed the S-methyltransferase TmtA which uses the same substrate as GliT but converts it stepwise to bis(methylthio)gliotoxin for regulatory purposes.
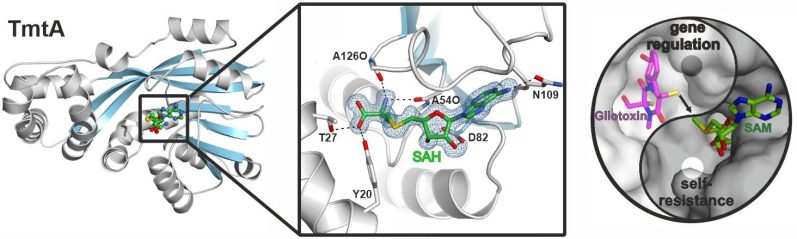
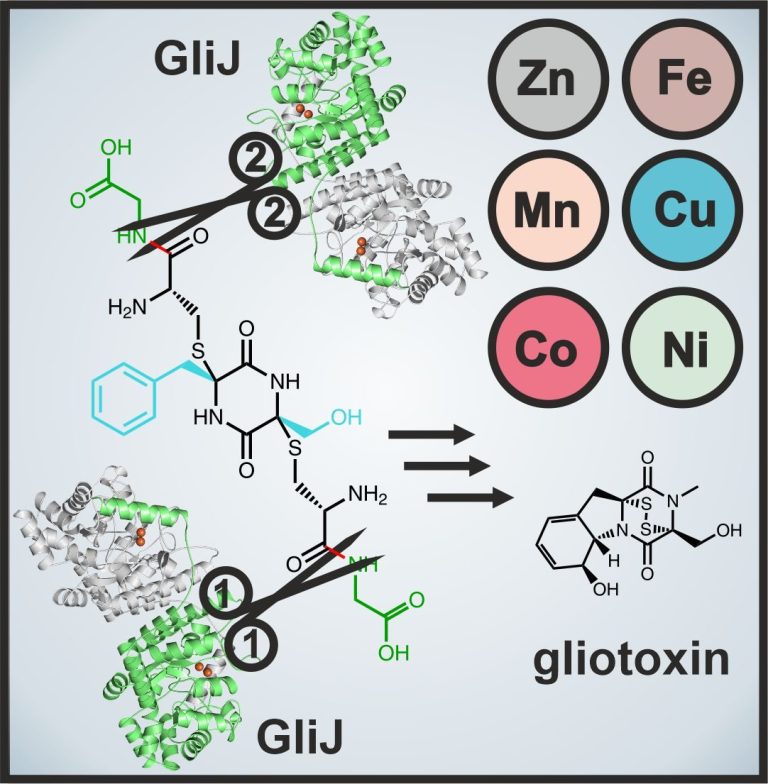
X-ray crystallography also proved a valuable method to study the dinuclear metallohydrolase GliJ. This carboxypeptidase garbles the glutathione moieties step by step and displays a surprising metal promiscuity. For this work we teamed up with experts in molecular modeling techniques to simulate the ligand at the enzyme's active site.
Marion, A., Groll, M., Scharf, D. H., Scherlach, K., Glaser, M., Sievers, H., Schuster, M., Hertweck, C., Brakhage, A. A., Antes, I., Huber, E. M.
Gliotoxin biosynthesis: Structure, mechanism and metal promiscuity of carboxypeptidase GliJ
ACS Chem. Biol., 2017, 12 (7), 1874-1882
Duell, E. R., Glaser, M., Le Chapelain, C., Antes, I., Groll, M., Huber, E. M.
Sequential inactivation of gliotoxin by the S-methyltransferase TmtA
ACS Chem. Biol., 2016, 11 (4), 1082–9
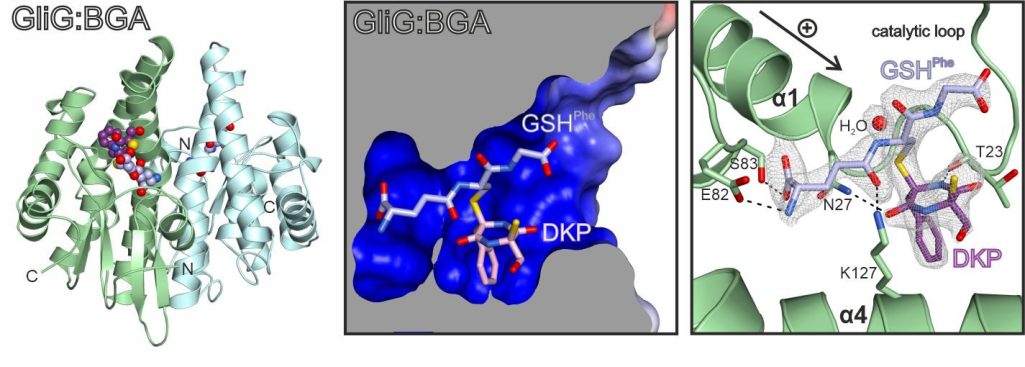
Most recently, we explored the mechanistic aspects of sulfur incorporation. A crystal structure of the glutathione-S-transferase GliG in complex with its reaction product, the bisglutathione adduct (BGA), suggests that the enzyme favors dehydration of the substrate and subsequently attaches glutathione. Stepwise processing of both substrate halves is required for full sulfurization.
Scherlach, K., Kuttenlochner, W., Scharf, D. H., Brakhage, A. A., Hertweck, C., Groll, M., Huber, E. M.
Structural and mechanistic insights into C-S bond formation in gliotoxin
Angew. Chem. Int. Ed., 2021, 60 (25), 14188-14194
Proteasome biology
The 20S proteasome is a barrel-shaped multicatalytic protease assembled from up to 14 different subunits. Due to its key role for intracellular protein degradation, activity of the 20S proteasome needs to be tightly controlled. To this end higher eukaryotes produce distinct proteasome regulators and proteasome types to fine-tune proteolytic activity. Moreover, they use sophisticated assembly lines to avoid premature, uncontrolled activity, and defects in this proteasome maturation sequence - similar as proteasome subunit overexpression - have been associated with inflammatory diseases. Proteasome inhibitors are therefore being investigated as anti-inflammatory agents, in addition to their well-established anti-cancer activity. Although the anti-proliferative effects of proteasome inhibitors are best studied in eukaryotes, microorganisms represent their largest repository. In fact, many microorganisms produce and secrete proteasome inhibitors in fight for their ecological niche or in response to environmental stimuli.
Using structural and biochemical techniques we address basic questions of proteasome assembly, maturation, activity, inhibition, drug resistances and evolution.
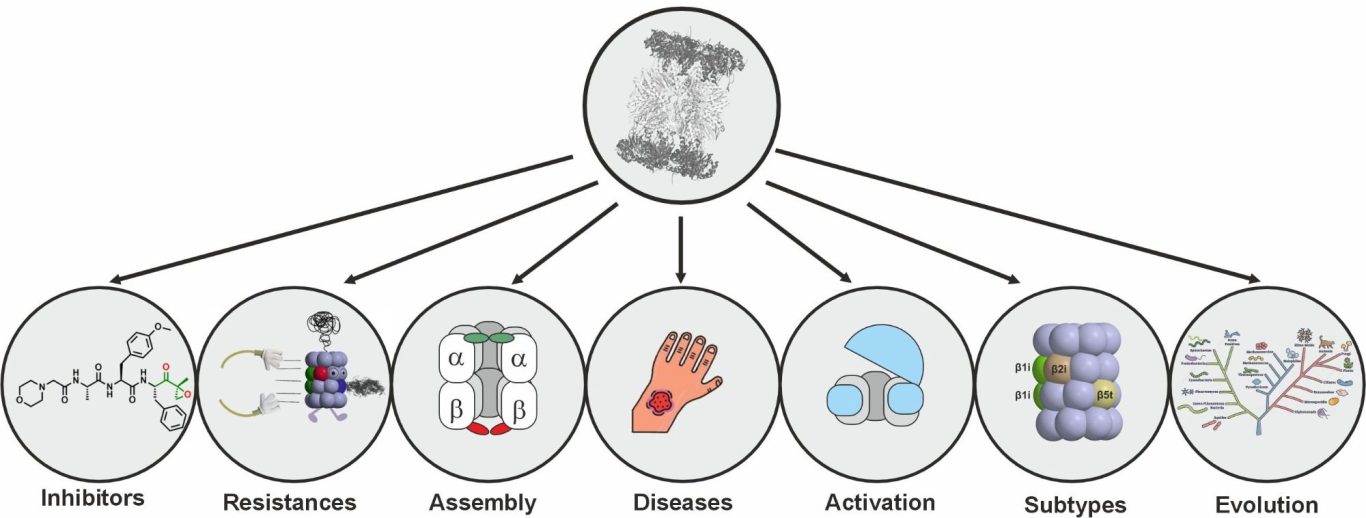
For more details on current and past proteasome projects, see our publication list.
Contact
PD Dr. Eva Maria Huber
Technical University of Munich
Center for Functional Protein Assemblies, room 02.037
Ernst-Otto-Fischer-Str. 8
85748 Garching b. München
Germany
Phone: +49-89-289-13354
E-Mail: eva.huber@tum.de

Funding

Hans-Fischer-Gesellschaft e. V.
Wir benötigen Ihre Zustimmung zum Laden der Übersetzungen
Wir nutzen einen Drittanbieter-Service, um den Inhalt der Website zu übersetzen, der möglicherweise Daten über Ihre Aktivitäten sammelt. Bitte überprüfen Sie die Details in der Datenschutzerklärung und akzeptieren Sie den Dienst, um die Übersetzungen zu sehen.